Classpath 0.91 is released with 1.45 million lines of code and with 98.96% coverage of Java 1.4.2, and 99.82% of java.swing. Or, as Dave calls it: 0.91 rocks! JChemPaint runs again (they fixed the XML parsing problem), and Jmol still runs , but slow. I also tested Taverna which now also starts up, but has an XML parsing error too: Exception occured whilst loading RDFS!
BioMed Central is setting up a new peer-reviewed, open access journal Source Code for Biology and Medicine. It will “encompass all aspects of workflow for information systems, decision support systems, client user networks, database management, and data mining” . Basically, anything that fits into chem-bla-ics. (Thanx to Werner, for pointing me to the website!) The ‘source code’ aspect is the interesting thing of this new journal.
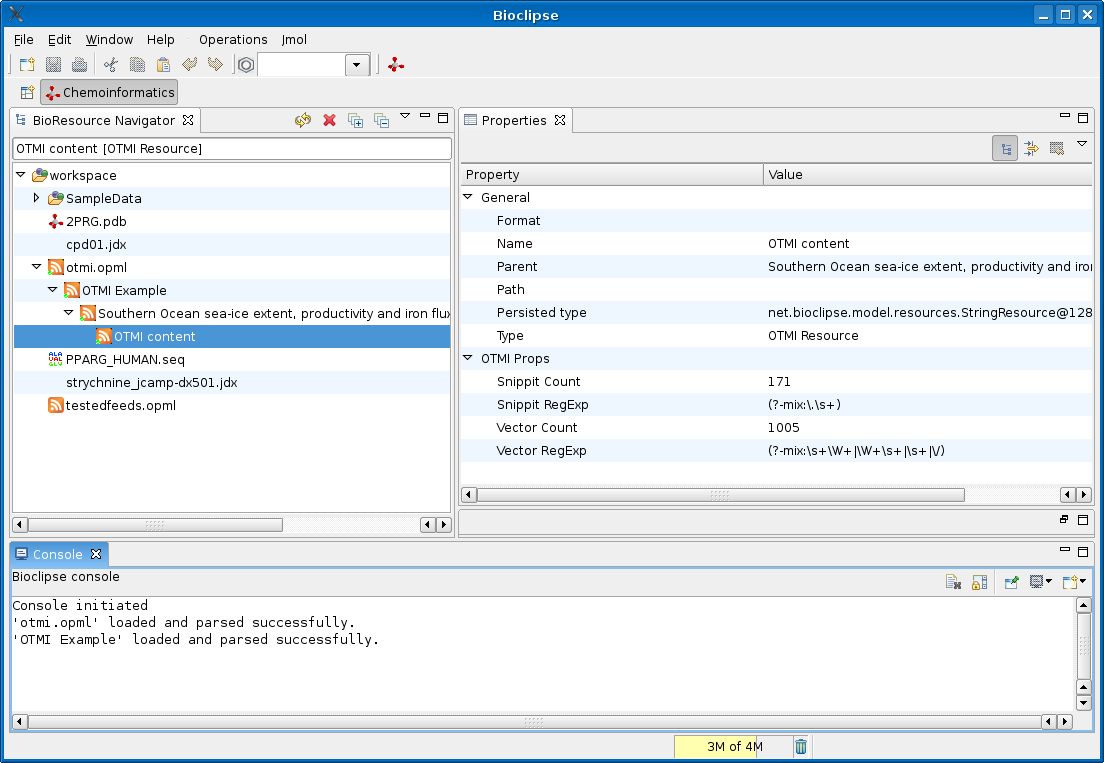
Timo Hannay blogged in Nature’s Nascent blog about the Open Text Mining Interface (OTMI), which is “a suggestion from Nature about how we might achieve text-mining and indexing purposes”. The idea is that each article has a link pointing to a machine readable file containing raw data about (and from?) the article.
Joerg Wegner mentioned in his blog the graph mining program ParMol which integrates four mining algorithms: MoSS (aka MoFa) and Gaston, which I mentioned in November last year , and FFSM and gSpan, which I did not know about yet. ParMol provides a common interface to the four different algorithms and is, like the four mining modules, licensed GPL.
Rajarshi Guha has set a nightly build service for the Chemistry Development Kit (CDK). The output is pretty, but information rich: it includes results for the JUnit test, DocCheck, and PMD. The compiled jar and the corresponding JavaDoc can be downloaded, offering a cutting edge distribution for users.
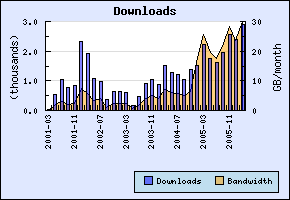
Here are some quick download statistics for some of the chemblaics components. First Jmol. The new stable Jmol 10.2 was release just over a week ago, and this obviously boosted downloads, breaking the monthly download total of two earlier this year (source): Statistics for the CDK include download numbers for the CDK library itself, but for JChemPaint, the CDK News, and several other packages too. Totals are at about 1/3rd of Jmol.
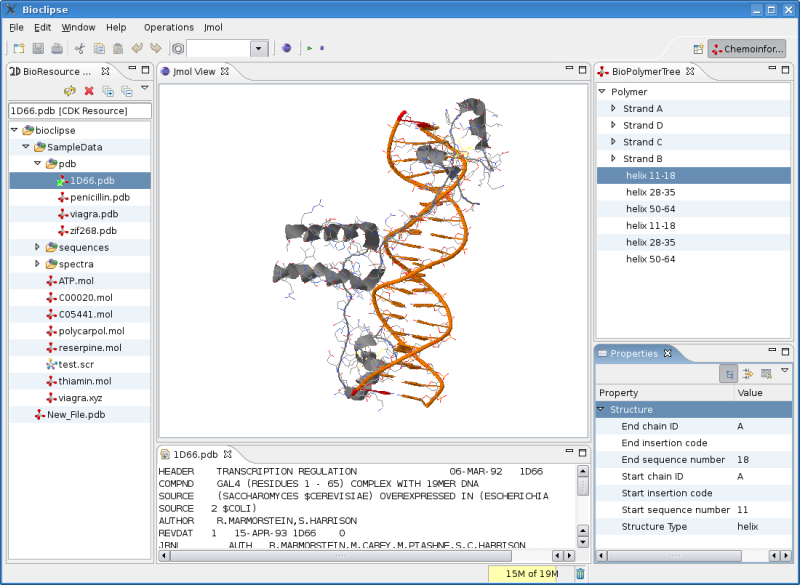
I have not blogged for about a week now, and been too busy with other things, like finishing my PhD articles/manuscript, my new job at the CUBIC where I continued the work on proper protein support in Bioclipse using the CDK and Jmol: The latter involves getting the CdkJmolAdapter , the interface between the CDK and Jmol, updated for changes since the Jmol as 3D viewer for CDK article in CDK News , the open access journal for CDK related
Conference season is nearing. And just in time, Postgenomic.com added a conferences map showing locations of upcoming and recently finished conferences. Oh boy, do I want to set this up for chemoinformatics too! Postgenomic.com makes use of the rel=“conference” attribute for the element.
The data classes of the Chemistry Development Kit are mutable, unlike those of Octet. This means that other classes may need to respond when the content updates. For example, a render class. CDK’s ChemObject provides a notifyChanged() and addListener() methods for this.
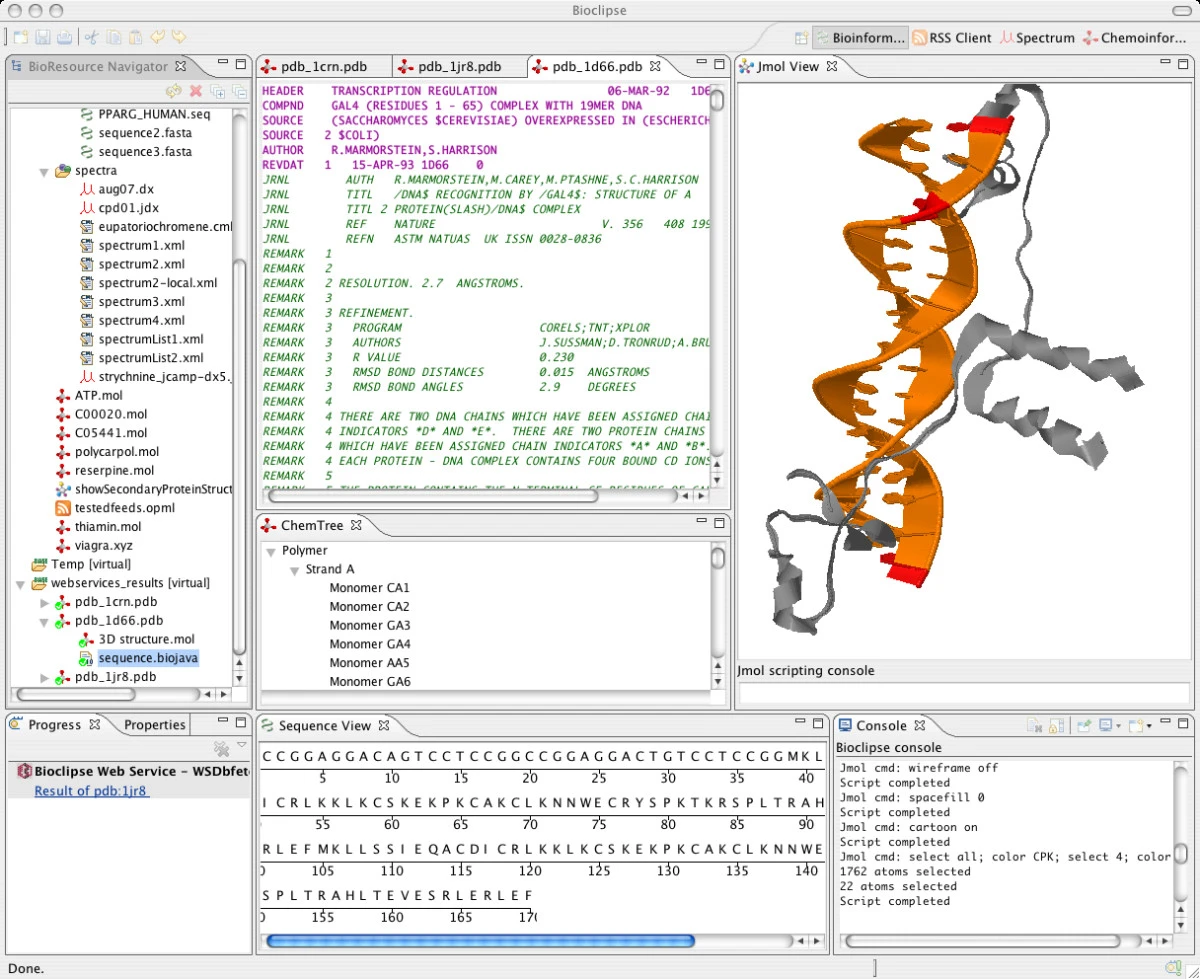
Bioclipse 1.0 is to be released in May, and the cartoon on script command is still not working in the Jmol viewer. For those who do not know yet, Bioclipse is a cool Eclipse RCP based Java chemo-and bioinformatics workbench. To have a better idea what goes on inside Bioclipse, I wrote a new BioPolymer tree to show me the strands in the protein.