I've been experimenting with simple spatial search in BioStor, as shown in the demo below. If you go to the map on BioStor you can use the tools on the left to draw a box or a polygon on the map, and BioStor will search it's database for articles that mention localities that occur in that region. If you click on a marker you can see the title of the article, clicking on that title takes you to the article itself.
iPhylo

I will be at ReCon 16 in Edinburgh (hashtag #ReCon_16), the second ReCon event I've attended (see Thoughts on ReCon 15: DOIs, GitHub, ORCID, altmetric, and transitive credit). For the hack day that follows I've put together some instructions for a way to glue together annotations made by multiple people using hypothes.is. It works by using IFTTT to read a user's annotation stream (i.e., the annotations they've made) and then post those to a

OK, so the title is pure click bait, but here's the thing. It seems to me that the Semantic Web as classically conceived (RDF/XML, SPARQL, triple stores) has had relatively little impact outside academia, whereas other technologies such as JSON, NoSQL (e.g., MongoDB, CouchDB) and graph databases (e.g., Neo4J) have got a lot of developer mindshare. In biodiversity informatics the Semantic Web has been a round for a while.

One of the limitations of the Biodiversity Heritage Library (BHL) is that, unlike say Google Books, its search functions are limited to searching metadata (e.g., book and article titles) and taxonomic names. It doesn't support full-text search, by which I mean you can't just type in the name of a locality, specimen code, or a phrase and expect to get back much in the way of results.
One of my pet projects is BioStor, which has been running since 2009 (gulp). BioStor extracts articles from the Biodiversity Heritage Library (details here: http://dx.doi.org/10.1186/1471-2105-12-187), and currently has over 110,000 articles, all open access. The site itself is showing its age, both in terms of performance and design, so I've wanted to update it for a while now.

My BioNames project has been going for over a year now, but I hadn't gotten around to providing bulk access to the data I've been collecting and cleaning. I've gone some way towards fixing this. You can now grab a snapshot of the BioNames database as a Darwin Core Archive here.
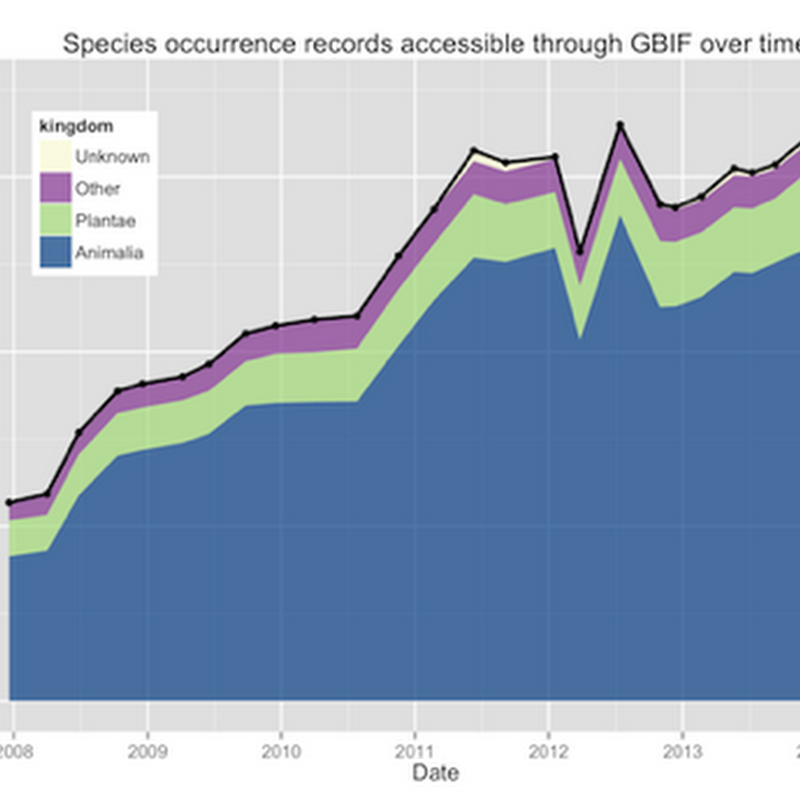
Tim Roberston and the ream at GBIF are working on some nice visualisations of GBIF data, and have made an early release available for viewing: http://analytics.gbif-uat.org.
Quick note to self about possible way to using fuzzy matching when searching for taxonomic names. Now that I'm using Cloudant to host CouchDB databases (e.g., see BioStor in the the cloud) I'd like to have a way to support fuzzy matching so that if I type in a name and misspelt it, there's a reasonable chance I will still find that name. This is the "did you mean?" feature beloved by Google users.

Quick note on an experimental version of BioStor that is (mostly) hosted in the cloud. BioStor currently runs on a Mac Mini and uses MySQL as the database. For a number of reasons (it's running on a Mac Mini and my knowledge of optimising MySQL is limited) BioStor is struggling a bit. It's also gathered a lot of cruff as I've worked on ways to map article citations to the rather messy metadata in BHL.

CrossRef have released CrossRef Metadata Search a nice tool that can take a free-form citation and return possible matches from CrossRef's database. If you get a match CrossRef can take the DOI and format for you it in a variety of styles using DOI content negotiation. If, like me, you spend a lot of time trying to find DOIs (and other identifiers) for articles by first parsing citations into their component parts, then this is good news.