Notes on how many georeferenced DNA sequences there are in GenBank, and how many could potentially be georeferenced.
iPhylo

Note to self for upcoming discussion with JournalMap. As of Monday August 25th, BioStor has 106,617 articles comprising 1,484,050 BHL pages. From the full text for these articles, I have extracted 45,452 distinct localities (i.e., geotagged with latitude and longitude). 15,860 BHL pages in BioStor pages have at least one geotag, these pages belong to 5,675 BioStor articles. In summary, BioStor has 5,675 full-text articles that are geotagged.

Following on from earlier posts on annotating biodiversity data (Rethinking annotating biodiversity data and More on annotating biodiversity data: beyond sticky notes and wikis) I've started playing with user interfaces for editing data.
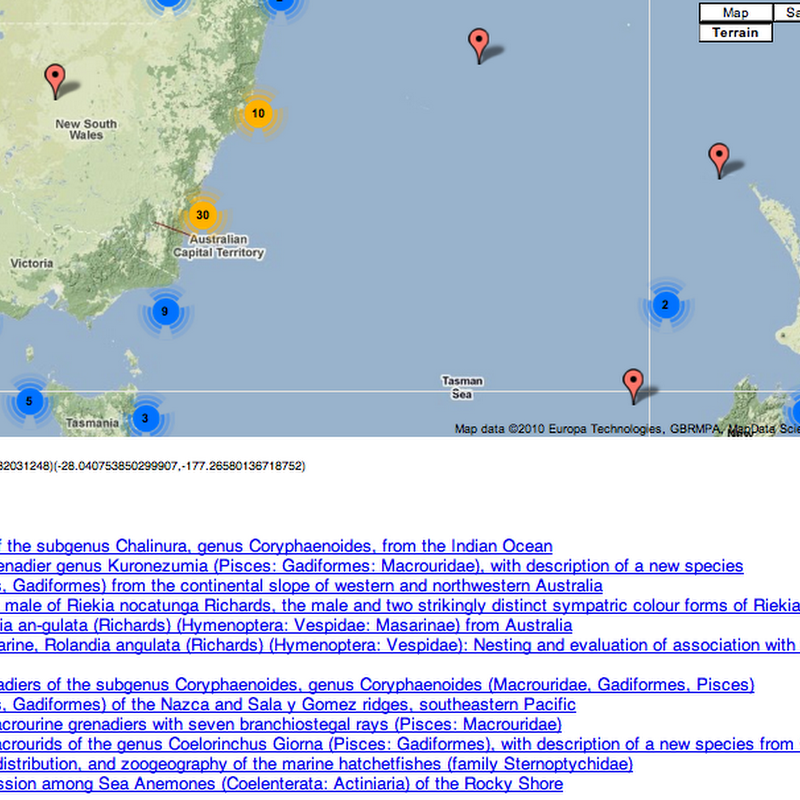
Every so often I revisit the idea of browsing a collection of documents (or specimens, or phylogenies) geographically. It's one thing to display a map of localities for single document (as I did most recently for Zootaxa ), it's quite another to browse a large collection.

I'm in the midst of rebuilding iSpecies (my mash-up of Wikipedia, NCBI, GBIF, Yahoo, and Google search results) with the aim of outputting the results in RDF. The goal is to convert iSpecies from a pretty crude "on-the-fly" mash-up to a triple store where results are cached and can be queried in interesting ways. Why?
Following on from my previous post about Wikispecies (which generated some discussion on TAXACOM) I've played some more with Wikispecies. AS a first step I've added a Wikispecies RSS feed to my list of RSS feeds. This feed takes the original Wikispecies RSS feed for new pages (generated by the page Special:NewPages ) and tries to extract some details before reformatting it as an ATOM feed.

e-Biosphere '09 kicks off next week, and features the challenge: Originally I planned to enter the wiki project I've been working on for a while, but time was running out and the deadline was too ambitious. Hence, I switched to thinking about RSS feeds. The idea was to first create a set of RSS feeds for sources that lack them, which I've been doing over at http://bioguid.info/rss, then integrate these feeds in a useful way.

One advantage of flying to the US is the chance to do some reading. At Newark (EWR) I picked up Guy Kawasaki's "Reality Check", which is a fun read. You can get a flavour of the book from this presentation Guy gave in 2006. While at MIT for the Elsevier Challenge I was browsing in the MIT book shop and stumbled across "Google and the Myth of Universal Knowledge" by Frenchman Jean-Noël Jeanneney. It's, um, very French.
D. Ross Robertson has published a paper entitled "Global biogeographical data bases on marine fishes: caveat emptor" (doi:10.1111/j.1472-4642.2008.00519.x - DOI is broken, you can get the article here). The paper concludes: As I've noted elsewhere on this blog, and as demonstrated by Yesson et al.'s paper on legume records in GBIF (doi:10.1371/journal.pone.0001124) (not cited by Robertson), there are major problems with geographical information

The more I play with GBIF the more I come across some spectacular errors. Here's one small example of what can go wrong, and how easy it would be to fix at least some of the errors in GBIF. This is topical given that the recent review of EOL highlighted the importance of vetting and cleaning data.