Update: Angelique Hjarding and her co-authors have responded in a guest post on iPhylo. The quality and fitness for use of GBIF-mobilised data is a topic of interest to anyone that uses GBIF data.
iPhylo
I stumbled across this paper (found on the GBIF Public Library): The first sentence of the abstract makes the paper sound a bit of a slog to read, but actually it's a great fun, full of pithy comments on the state of digital humanities. Almost all of this is highly relevant to mobilising natural history data.
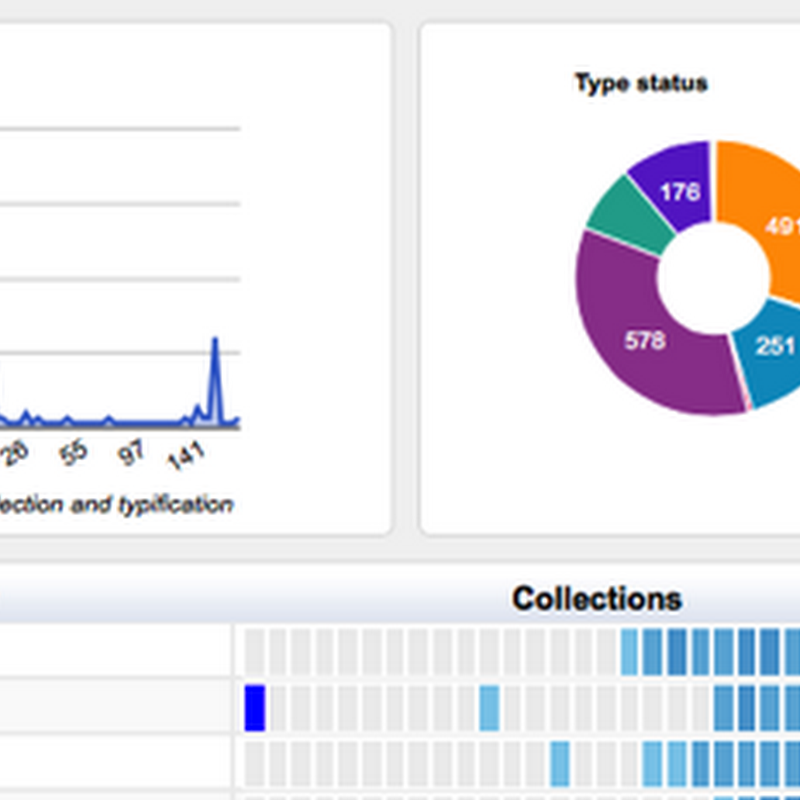
I'm adding more charts to the GBIF Chart tool, including some to explore the type status of specimens from the Solomon Islands. There are nearly 500 holotypes from this region, so quite a few new species have been discovered in this region.
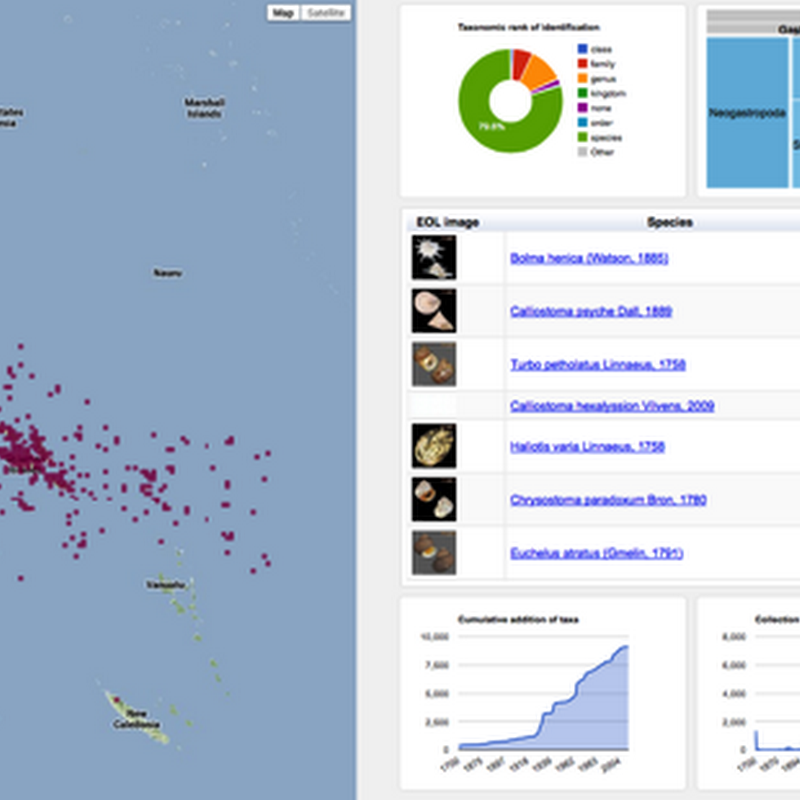
Following on from the previous post on visualising GBIF data, I've added some more interactivity.
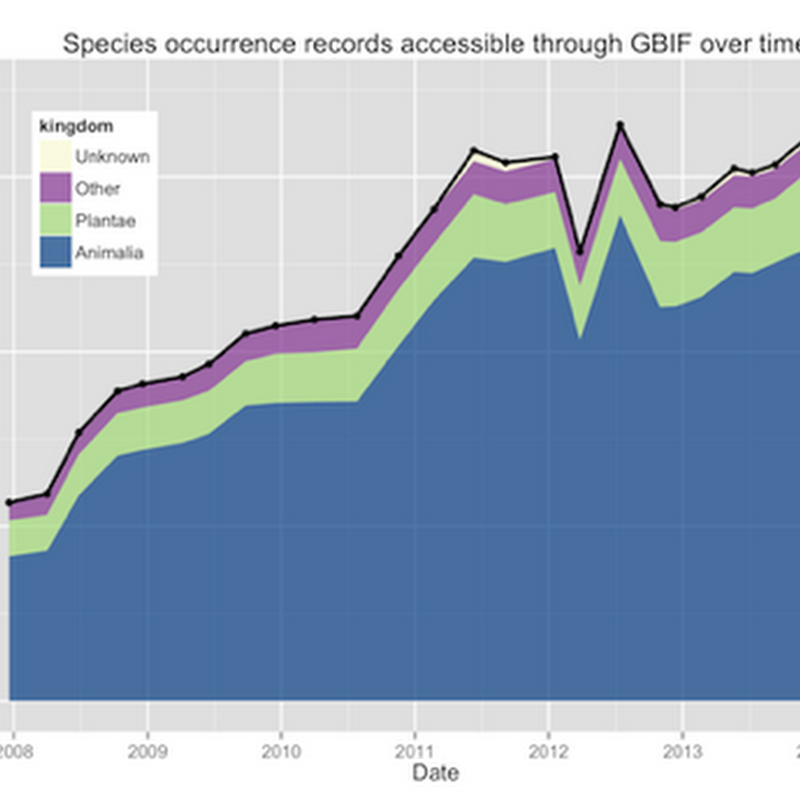
Tim Roberston and the ream at GBIF are working on some nice visualisations of GBIF data, and have made an early release available for viewing: http://analytics.gbif-uat.org.

Today I managed to publish some data from a GitHub repository directly to GBIF. Within a few minutes (and with Tim Robertson on hand via Skype to debug a few glitches) the data was automatically indexed by GBIF and its maps updated. You can see the data I uploaded here. The data I uploaded came from this paper: This is the data I used to build the geophylogeny for Banza using Google Earth.

As part of a project exploring GBIF data I've been playing with displaying GBIF data on Google Maps.

A quick note to myself to document a problem with the GBIF classification of liverworts (I've created issue POR-1879 for this). While building a new tool to browse GBIF data I ran into a problem that the taxon "Jungermanniales" popped up in two different places in the GBIF classification, which broke a graphical display widget I was using.
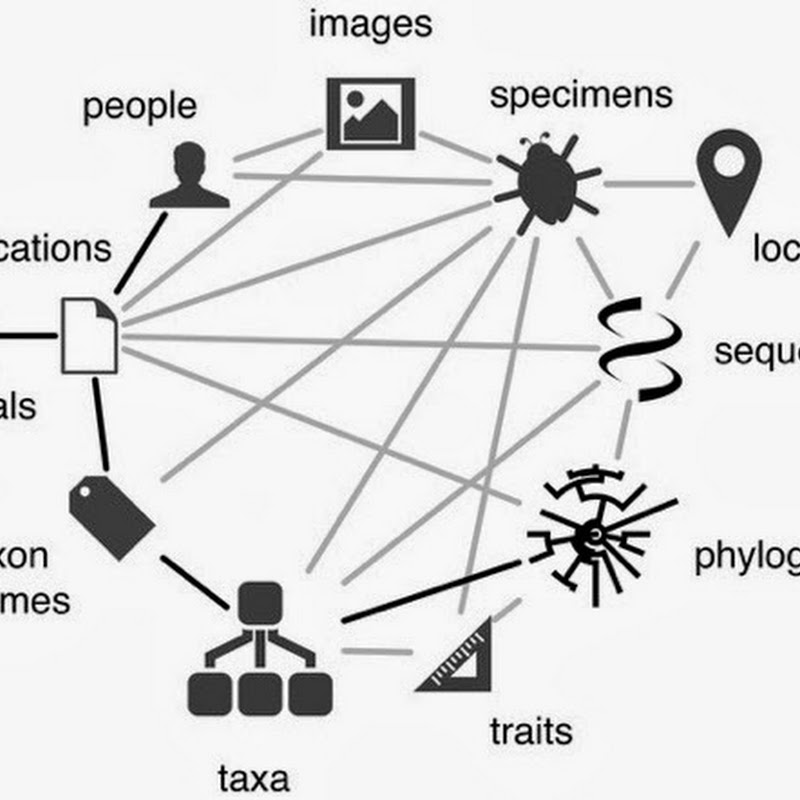
More for my own benefit than anything else I've decided to list some of the things I plan to work on this year. If nothing else, it may make sobering reading this time next year. A knowledge graph for biodiversity Google's introduction of the "knowledge graph" gives us a happy phrase to use when talking about linking stuff together.
Given that it's the start of a new year, and I have a short window before teaching kicks off in earnest (and I have to revise my phyloinformatics course) I'm playing with a few GBIF-related ideas. One topic which comes up a lot is annotating and correcting errors. There has been some work in this area [1][2] bit it strikes me as somewhat complicated. I'm wondering whether we couldn't try and keep things simple.