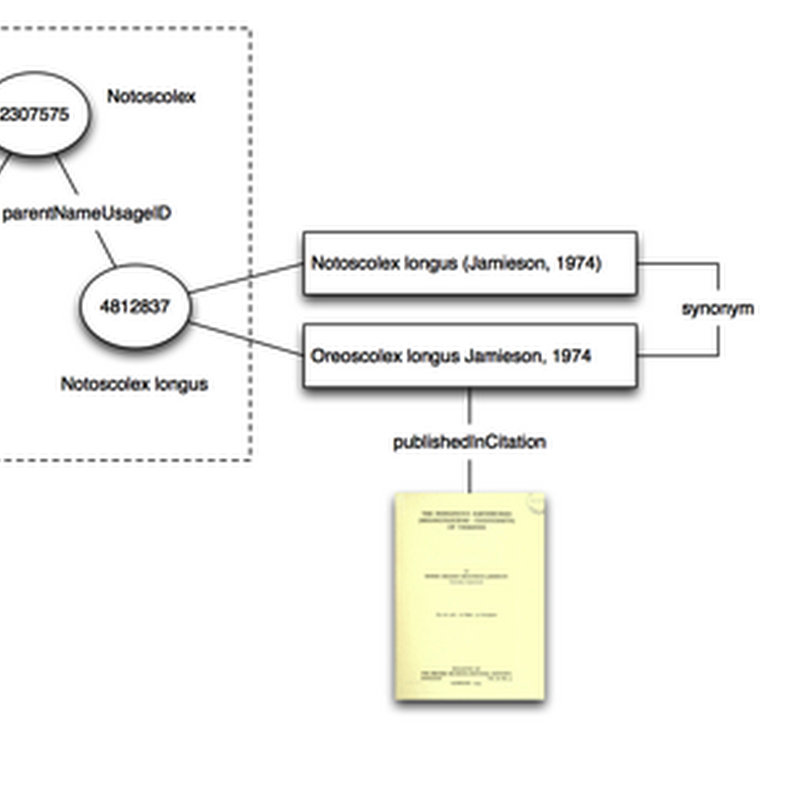
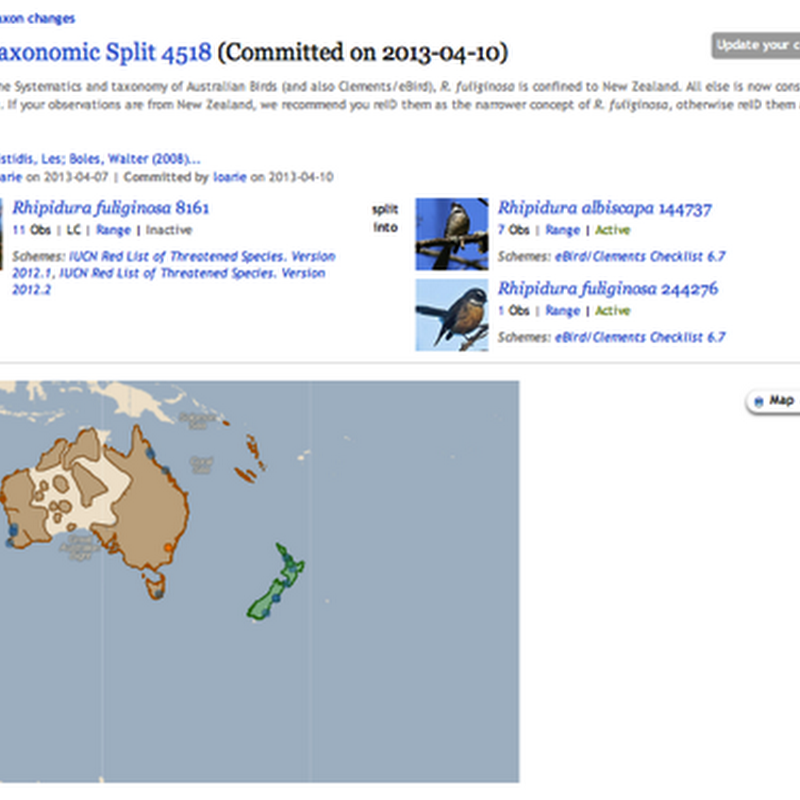
One reason I built BioNames (and the related digital archive BioStor) was to create tools to help make sense of taxonomic names. In exploring databases such as GBIF and the NCBI taxonomy every so often you come across cases where things have gone horribly wrong, and to make sense of them you have to drill down into the taxonomic literature.