Editor’s note: This is the first in a series of posts from rOpenSci’s recent hackathon. I recently had the pleasure of participating in rOpenSci’s hackathon. To be honest, I was quite nervous to work among such notables, but I immediately felt welcome thanks to a warm and personable group. Alyssa Frazee has a great post summarizing the event, so check that out if you haven’t already.
We’ve received a number of questions from our users about dealing with the finer details of data sources on the web. Whether you’re reading data from local storage such as a csv file, a .Rdata store, or possibly a proprietary file format, you’ve most likely run into some issues in the past. Common problems include passing incorrect paths, files being too big for memory, or requiring several packages to read files in incompatible formats.
The iNaturalist project is a really cool way to both engage people in citizen science and collect species occurrence data. The premise is pretty simple, users download an app for their smartphone, and then can easily geo reference any specimen they see, uploading it to the iNaturalist website. It let’s users turn casual observations into meaningful crowdsourced species occurrence data.
UPDATE: mapping functions are in a separate package now (mapr). Examples that do mapping below have been updated. The rOpenSci projects aims to provide programmatic access to scientific data repositories on the web. A vast majority of the packages in our current suite retrieve some form of biodiversity or taxonomic data.
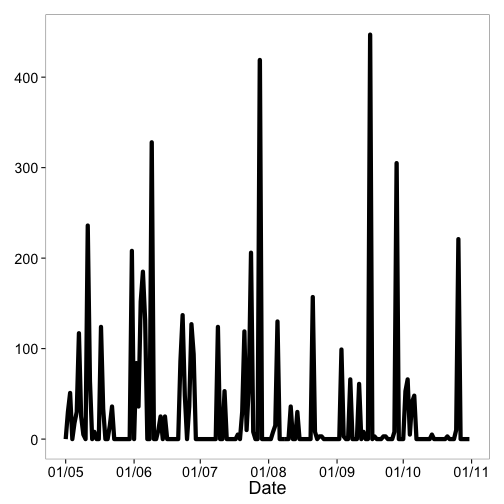
We recently pushed the first version of rnoaa to CRAN - version 0.1. NOAA has a lot of data, some of which is provided via the National Climatic Data Center, or NCDC. NOAA has provided access to NCDC climate data via a RESTful API - which is great because people like us can create clients for different programming languages to access their data programatically.
Reproducible research involves the careful, annotated preservation of data, analysis code, and associated files, such that statistical procedures, output, and published results can be directly and fully replicated. As the push for reproducible research has grown, the R community has responded with an increasingly large set of tools for engaging in reproducible research practices (see, for example, the ReproducibleResearch Task View on CRAN).

We just released a new version of taxize - version 0.2.0. This release contains a number of new features, and bug fixes. Here is a run down of some of the changes: First, install and load taxize install.packages("rgbif") library(taxize) New things New functions: class2tree Sometimes you just want to have a visual of the taxonomic relationships among taxa.

Data on more than 10,000 species of ants recorded worldwide are available through from California Academy of Sciences’ AntWeb, a repository that boasts a wealth of natural history data, digital images, and specimen records on ant species from a large community of museum curators.
rgbif is an R package to search and retrieve data from the Global Biodiverity Information Facilty (GBIF). rgbif wraps R code around the [GBIF API][gbifapi] to allow you to talk to GBIF from R. We just pushed a new verion of rgbif to cran - v0.5.0. Source and binary files are now available on CRAN. There are a few new functions: count_facet, elevation, and installations. These are described, with examples, below.