rOpenSci Software Peer Review’s guidance has been compiled in an online book for more than one year now. We’ve just released its fifth version.To find out what’s new in our dev guide 0.5.0, you can read the changelog,or this blog post for more digested information. A curation policy for rOpenSci packages The most exciting update to our guide is probably the addition of a chapter featuring rOpenSci package curation policy.
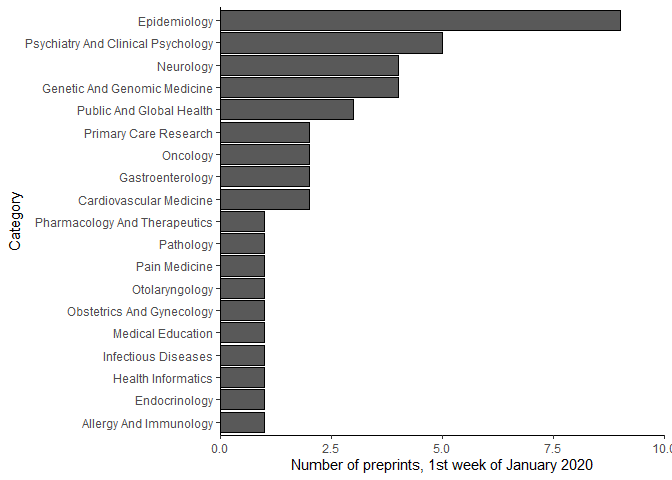
Background & motivation medRxiv, as the preprint repository for papers in the medical, clinical, and related health sciences, 1 has become a central source of new studies related to the COVID-19 pandemic.
mapscanner It is sometimes easy in the midst of the cutting-edge world of uniquesoftware development that is rOpenSci to forget that even though oursoftware might be freely available from anywhere in the world, access toadequate hardware is often restricted.
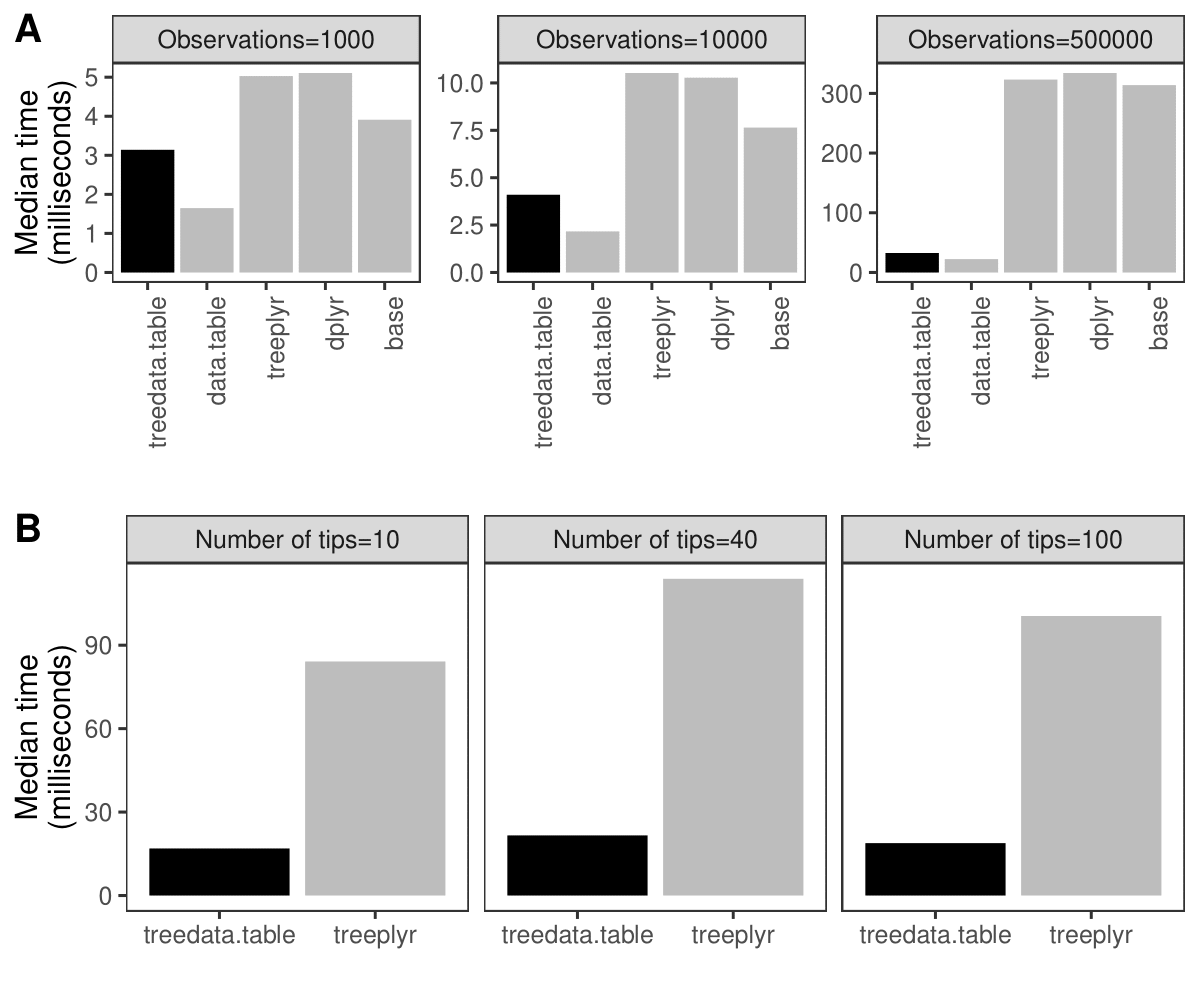
The data.table package enables high-performance extended functionality for data tables in R. treedata.table is a wrapperfor data.table for phylogenetic analyses that matches a phylogeny to the data.table, and preserves matching during data.table operations.Using the data.table package greatly increases analysis reproducibility and the efficiency of data manipulation operations over other ways of performing similar tasks inbase R, enabling

This post describes a few different aspects behind the scenes of the development of dittodb which recently went through the rOpenSci peer review process and was released to CRAN on 24 July 2020.This isn’t an introduction to the package itself (that’s available on dittodb’s site), but rather a look behind the scenes of the conceiving of the idea, the inspiration for, some of the development of, and history behind dittodb.
The R language has become very popular among scientists and analystsbecause it enables the rapid development of software and empowersscientific investigation. However, regardless of the language used,data analysis is usually complicated. Because of various projectcomplexities and time constraints, analytical software often reflectsthese challenges. “What did I measure? What analyses are relevant tothe study? Do I need to transform the data?
rOpenSci Software Peer Review’s guidance has been compiled in an online book for more than one year now. We’ve just released its fourth version.To find out what’s new in our dev guide 0.4.0, you can read the changelog,or this blog post for more digested information. Policy and governance changes Some aspects of the software review process changed.
parzer is a new package for handling messy geographic coordinates.The first version is now on CRAN, with binaries coming soon hopefully (seenote about installation below). The package recently completed rOpenScireview. parzer motivation The idea for this package started with a tweet from Noam Ross(https://twitter.com/noamross/status/1070733367522590721) about 15 months ago.
“Continuous Integration” (CI) has become a standard for proper software development.Checking the integrity of software after changes have been made is essential to ensure its proper functionality.Also, CI helps catch problems introduced by dependencies early when executed on a regular basis (usually done via scheduled CRON runs). Multiple professional providers exist (Travis CI, AppVeyor CI, Circle CI, etc.) which offer CI services to the
Dealing with taxonomic inconsistencies within and across datasets is a fundamental challenge of ecology and evolutionary biology. Accounting for species synonyms, taxa splitting and unification is especially important as aggregation of data across time and different data sources becomes increasingly common.