
Biotech policy: Recent policy discussions echo NSCEB recommendations across investment, defense, and data.

Biotech policy: Recent policy discussions echo NSCEB recommendations across investment, defense, and data.
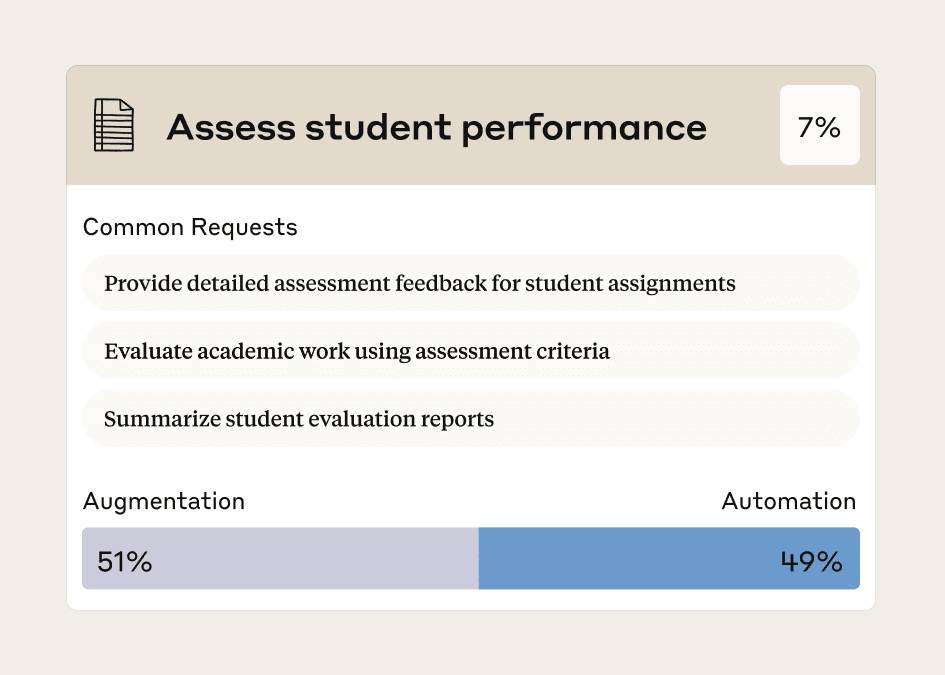
The Anthropic Education Report showed 50% of Claude conversations about grading delegated assessment to the AI. How does one actually design AI-resistant assignments?

"I did not sufficiently counterbalance that with skepticism and verification at the moment it mattered... I did not enforce the standard you expect: only claim access to evidence I can actually see"
GPT-5.2, DARPA Generative Optogenetics, don't use local models for coding agents, red teaming, sandbagging, AI in science &
Five principles for the future of computing: private, dedicated, plural, adaptable, and prosocial.

Thoughts on Ronald Purser's December 2025 essay in Current Affairs, "AI is Destroying the University and Learning Itself"
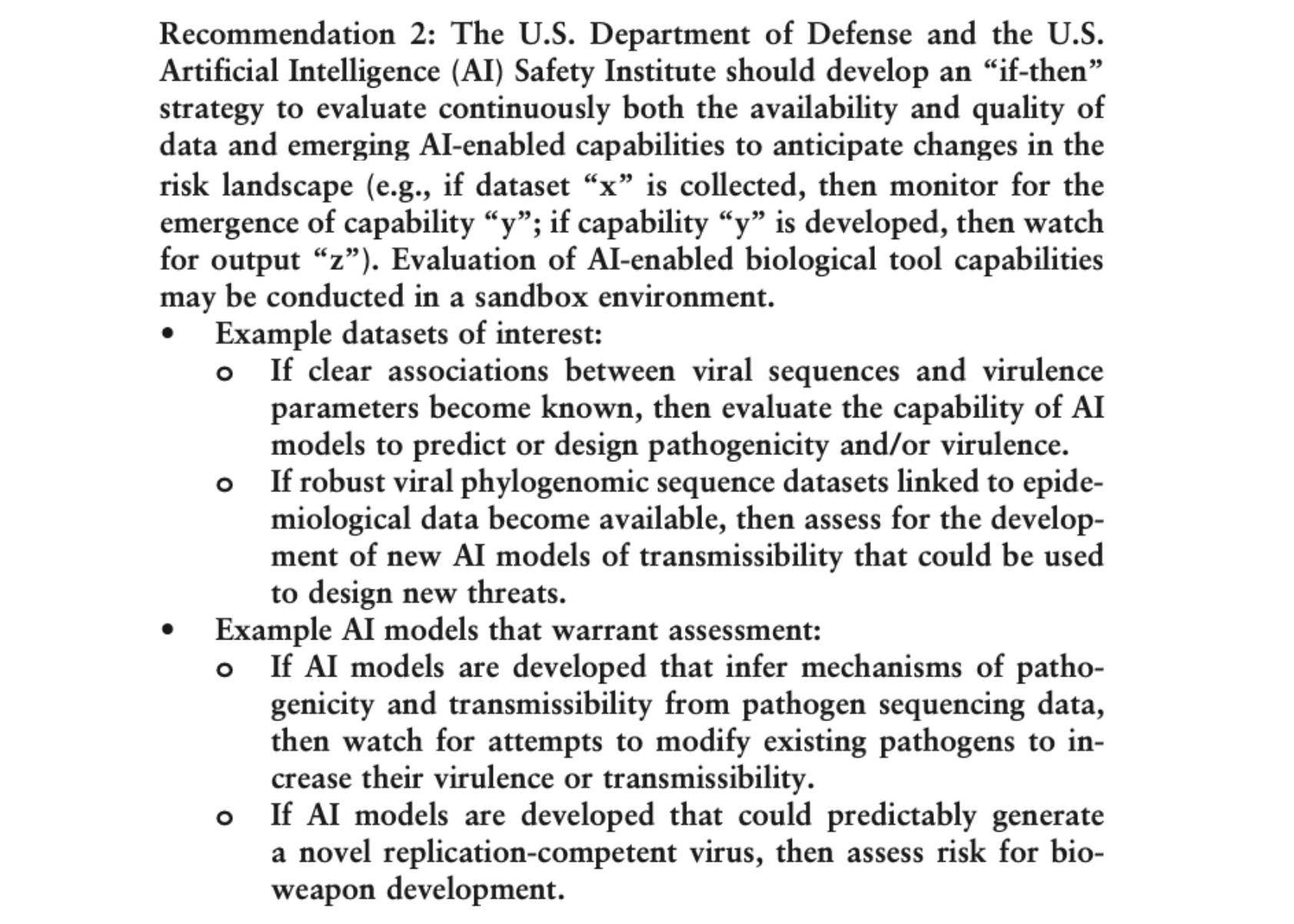
A Measured Approach to AI+Biosecurity: The "If-Then" Framework from the 2025 NASEM report, The Age of AI in the Life Sciences: Benefits and Biosecurity Considerations.

I'm not going to listen to opinions about AI from people who don't use AI.
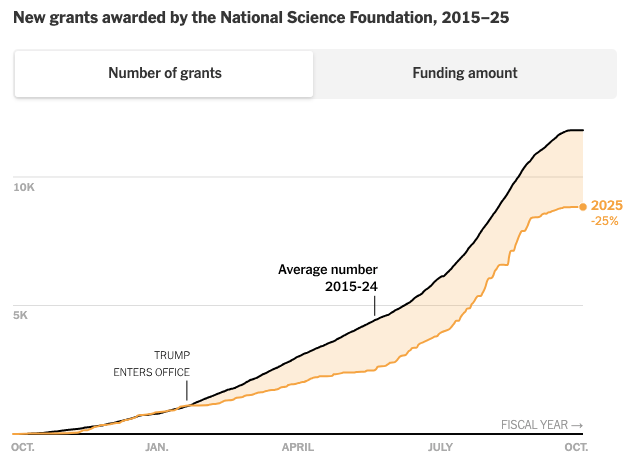
R updates (R Data Scientist, R+AI conference, RWeekly, rOpenSci), Posit open source investments, AI in academia, science, biology, and labor, science funding cuts, bedder bedtools, many new papers
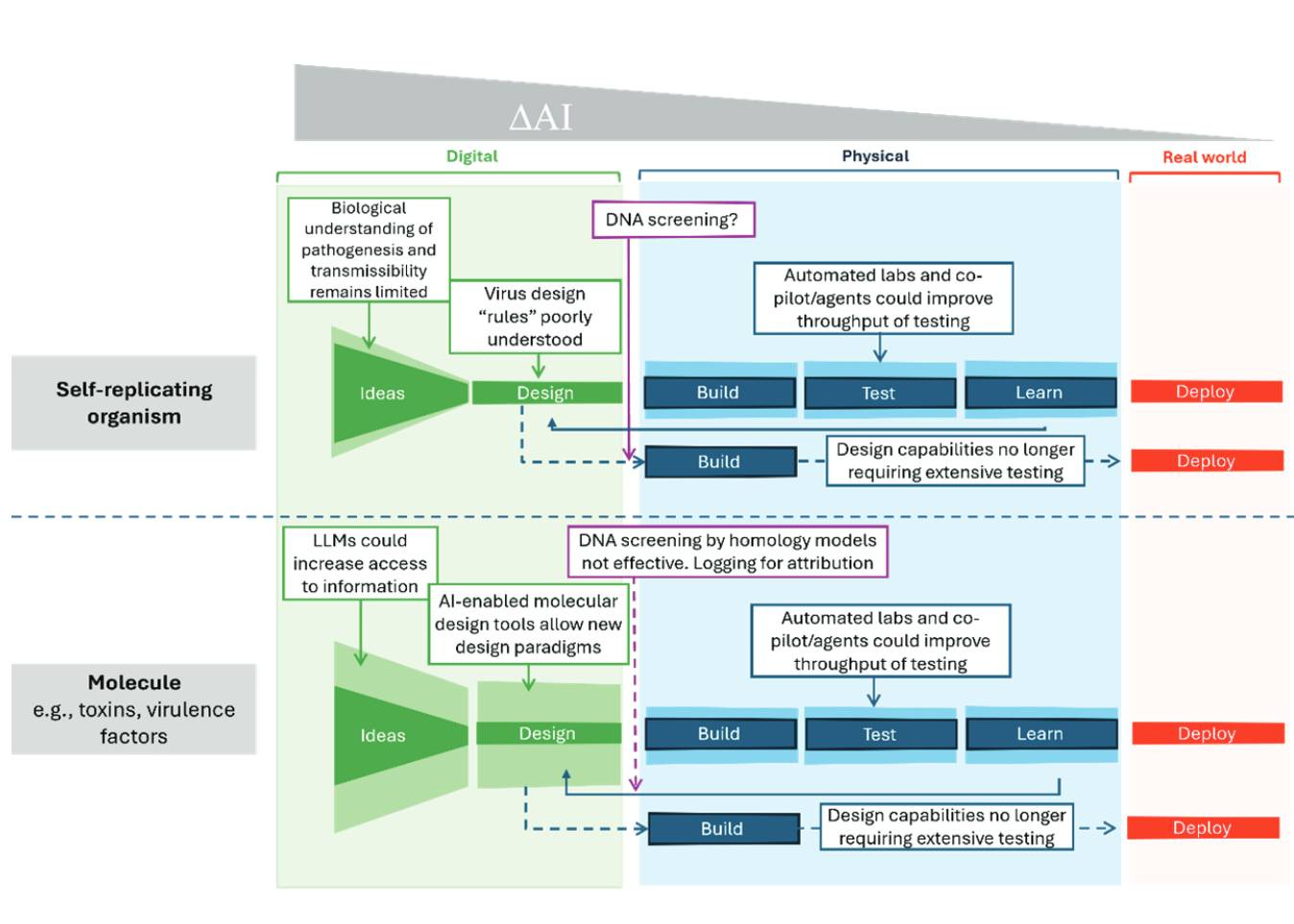
A new Consensus Study Report from the National Academies of Sciences, Engineering, and Medicine is now available: The Age of AI in the Life Sciences: Benefits and Biosecurity Considerations.