I recommend subscribing to Claus Wilke’s newsletter, Genes, Minds, Machines. I’ve linked to many of his essays in recent weeks. This one was a good read. Now that I’m back in academia and will be taking on Ph.D. students in my lab, this was good advice for me to read, as a future mentor.

In cybersecurity, defenders have to get everything right. Cyber defense must patch every vulnerability, secure every endpoint, anticipate every exploit. Attackers, by contrast, only need to find one overlooked flaw. That imbalance, which has shaped decades of cyber conflict, has a biological analogue.
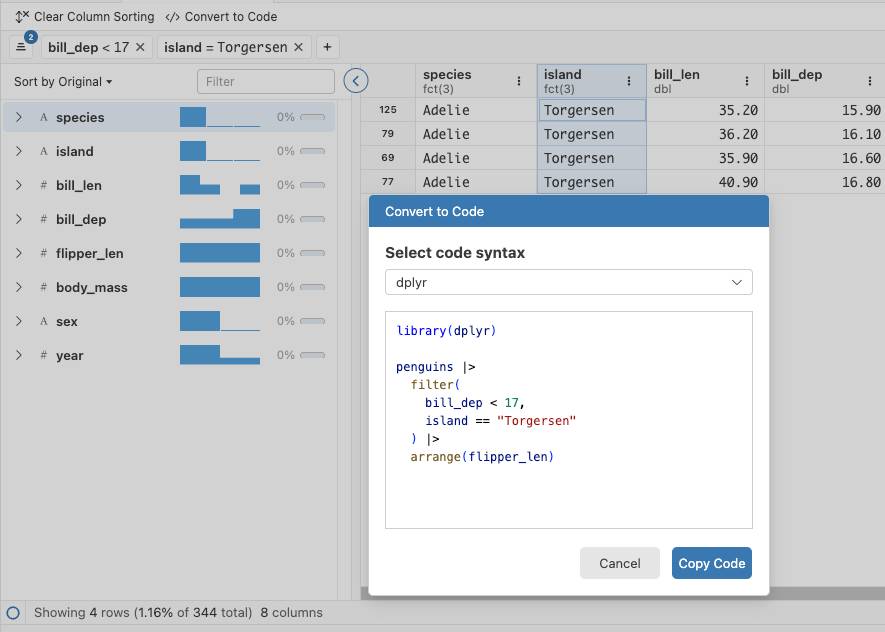
The most recent release of Positron (2025.10.0 build 199) has a few updates to the data explorer. Among other things, the actions you take in the data explorer like filtering and sorting can be converted to code: dplyr code for data frames, and SQL for DuckDB datasets.

Happy Friday, colleagues. It’s the end of the week and once again I’m going through my long list of idle browser tabs trying to catch up where I can. Lots of R and AI-related news this week. Subscribe now Strengthening nucleic acid biosecurity screening against generative protein design tools. This was a really cool paper published in Science this week from a team at Microsoft, Battelle, IDT, Twist, and others.
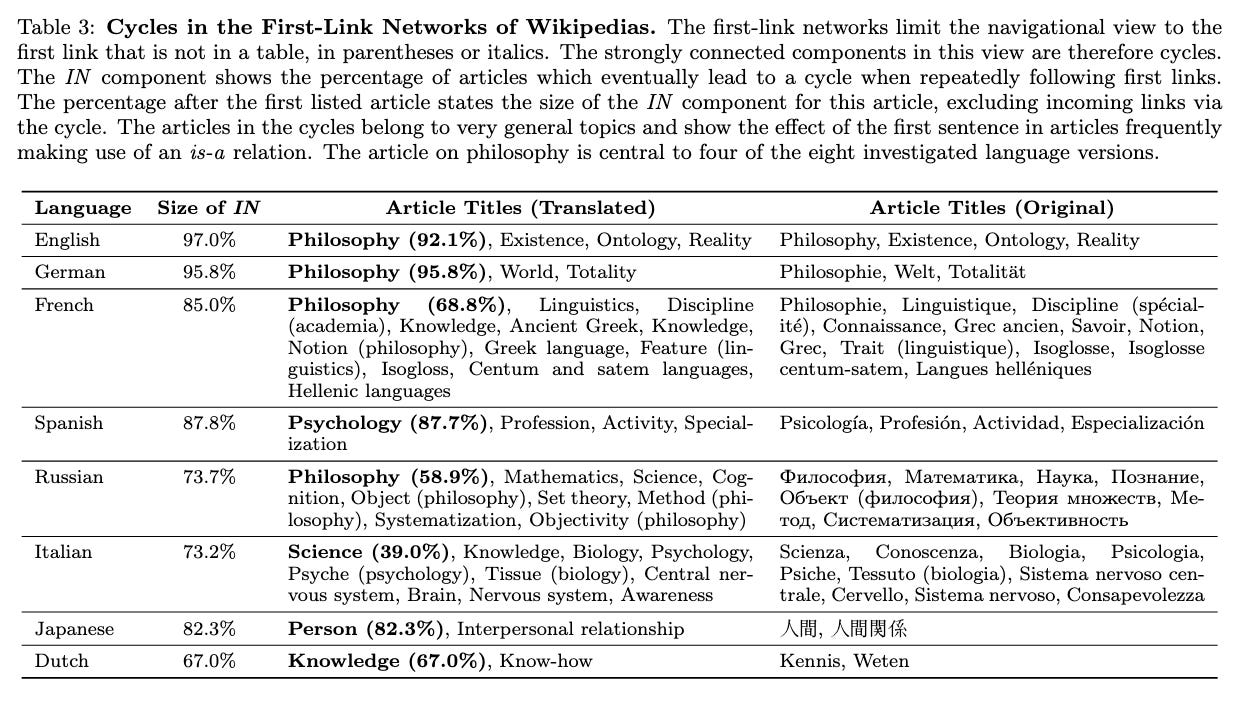
Over the weekend I stumbled across an interesting conference paper: Evaluating and Improving Navigability of Wikipedia . The article shows if you follow the first link in the main text of an English Wikipedia article, and then repeat the process for subsequent articles, it’ll lead you to Philosophy 97% of the time. I had my doubts, so I gave this a try with a few topics close to me. From Data science
Happy Friday, colleagues. It’s the end of the week and once again I’m going through my long list of idle browser tabs trying to catch up where I can. Lots of R and AI-related news this week. Subscribe now Emil Hvitfeldt: Slidecrafting (slidecrafting-book.com) . This is a really wonderful one-stop shop for tips on making beautiful slides with reveal.js and Quarto.
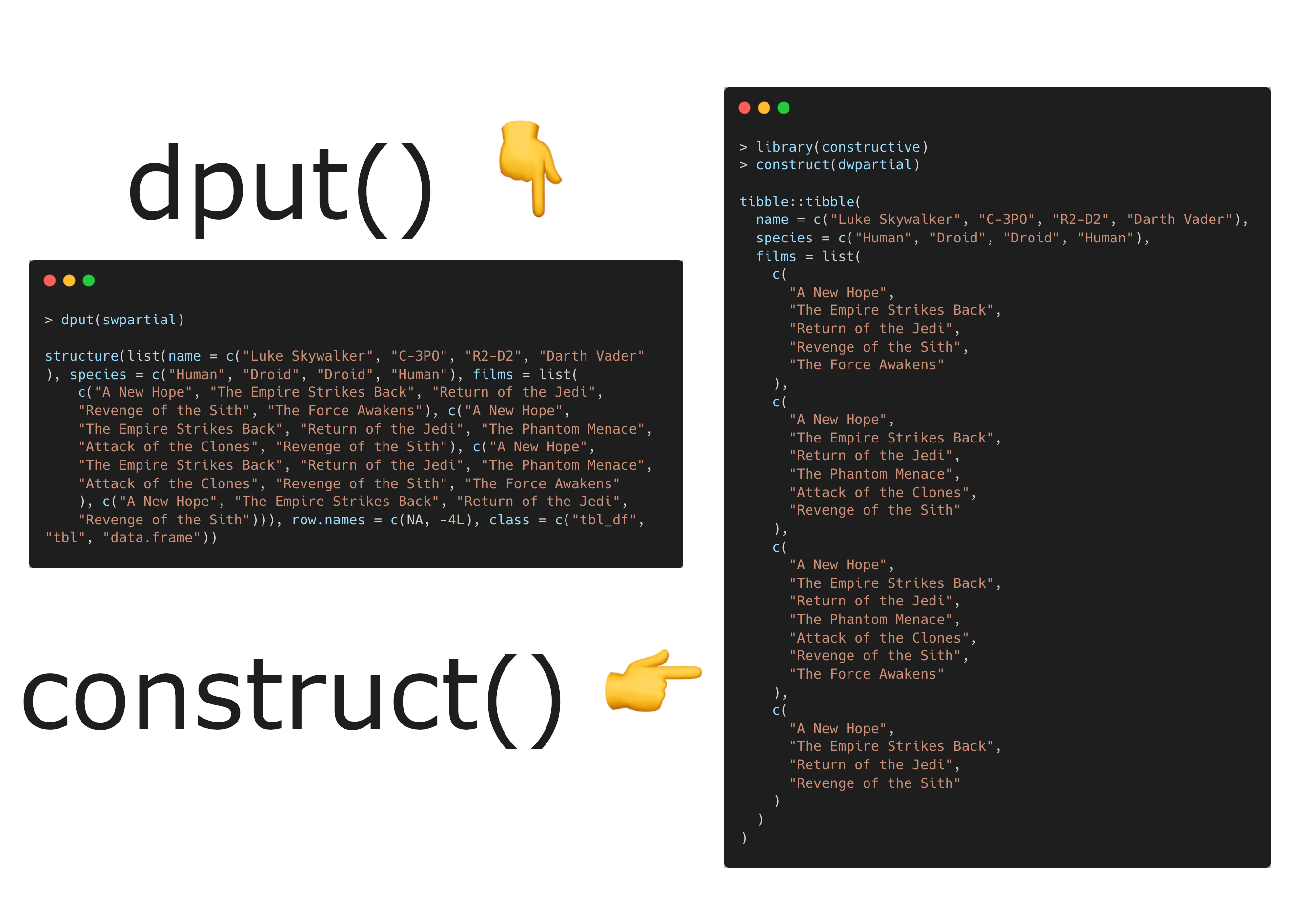
Today I discovered the constructive package and the construct() function for creating R objects with idiomatic R code to make human-readable reproducible examples. CRAN: https://cran.r-project.org/package=constructive Source: https://github.com/cynkra/constructive/ Docs &
Today I learned you can create and customize ggplot2 visualizations using your voice alone.
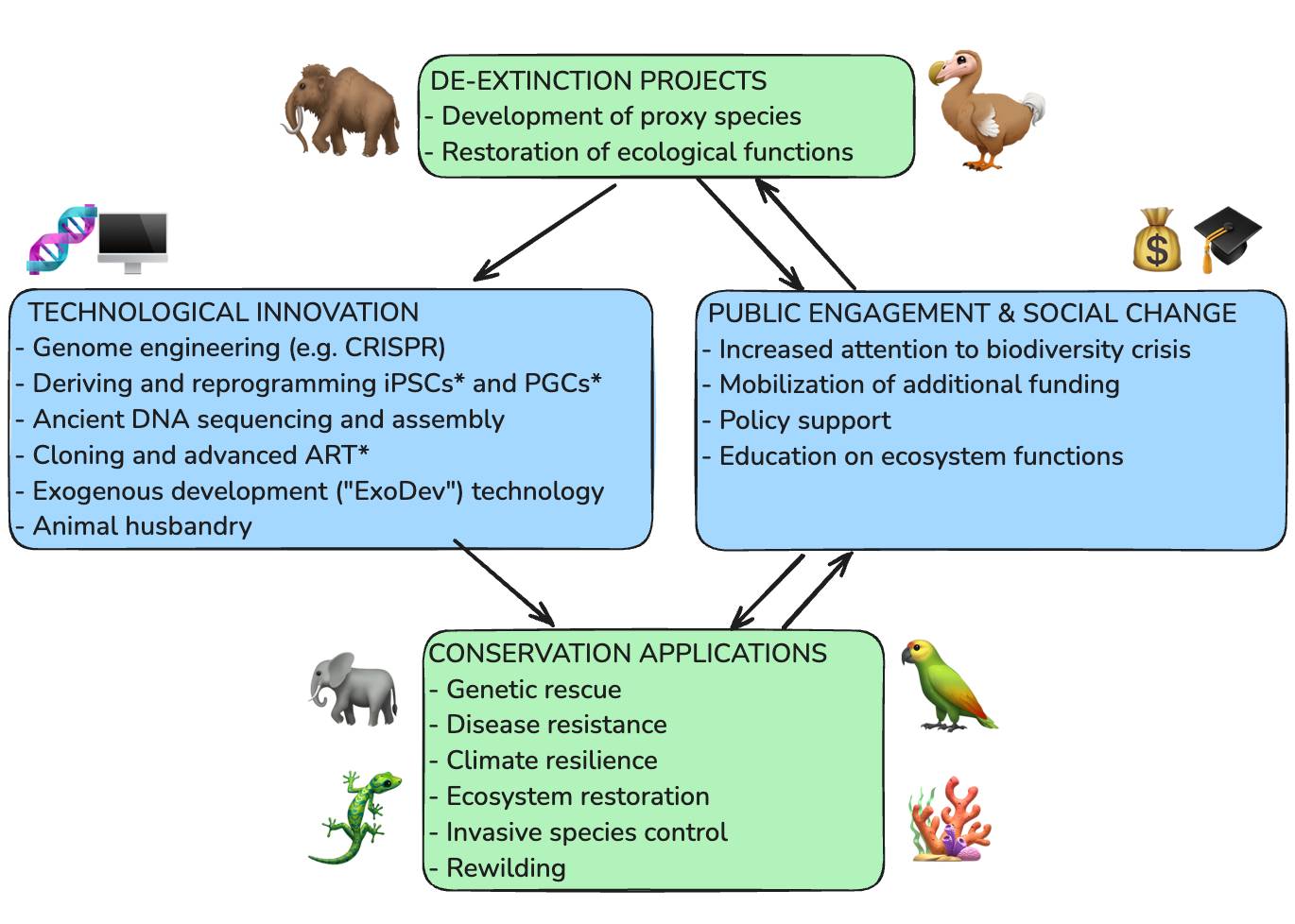
Happy Friday, colleagues. September has gone by at warp speed.

Last night we had our first event in the newly (re-)launched Charlottesville R Users (CRU) group. We had about 30 or so attendees — about half from academia and half from industry, with a few from local government organizations.
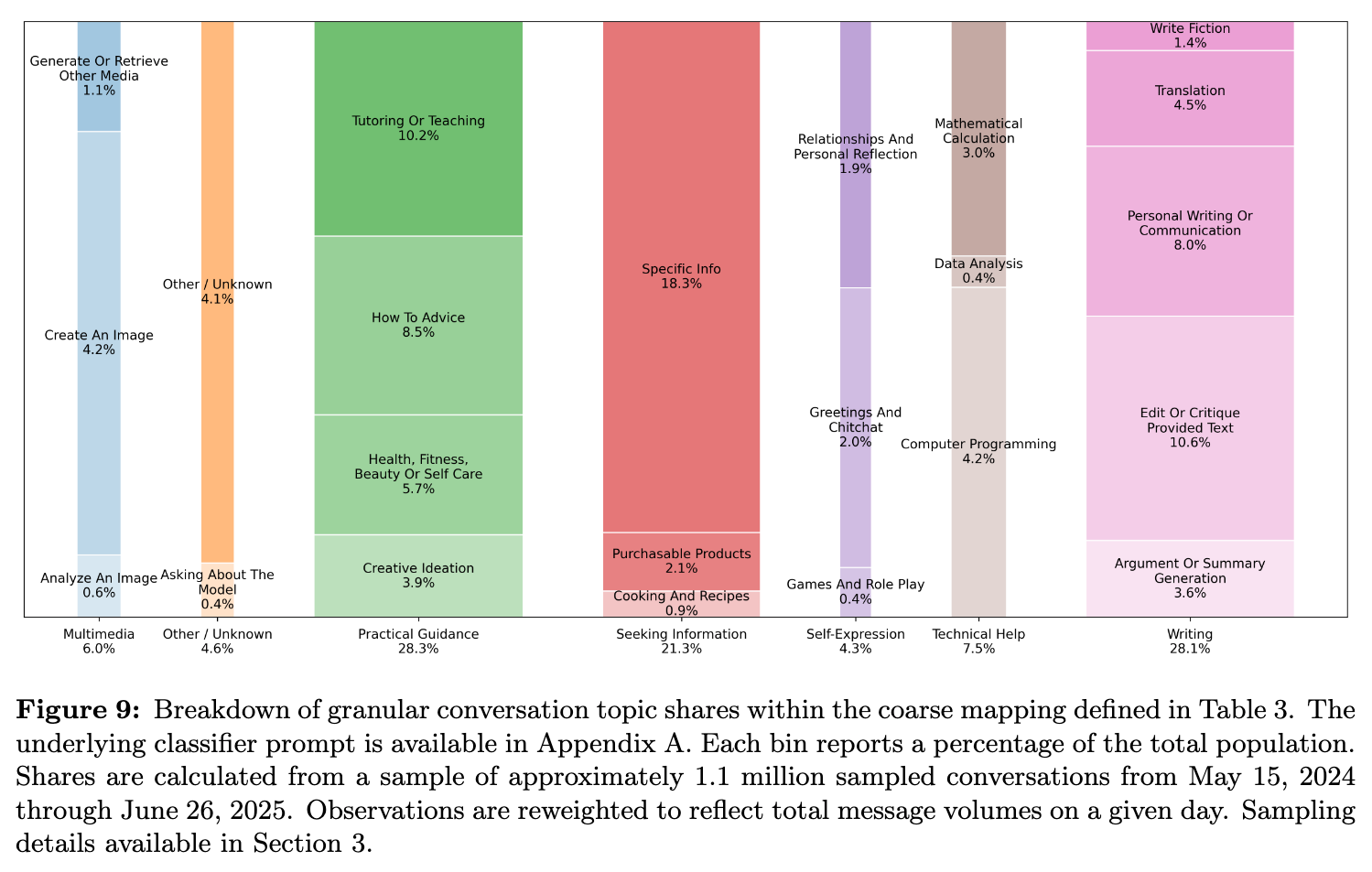
Happy Friday, colleagues. Lots going on this week. Once again I’m going through my long list of idle browser tabs trying to catch up where I can. ElevenReader (no affiliation) has been helpful to catch up this week! Subscribe now The Arc Institute reported the first ever viable genomes with genome language models.