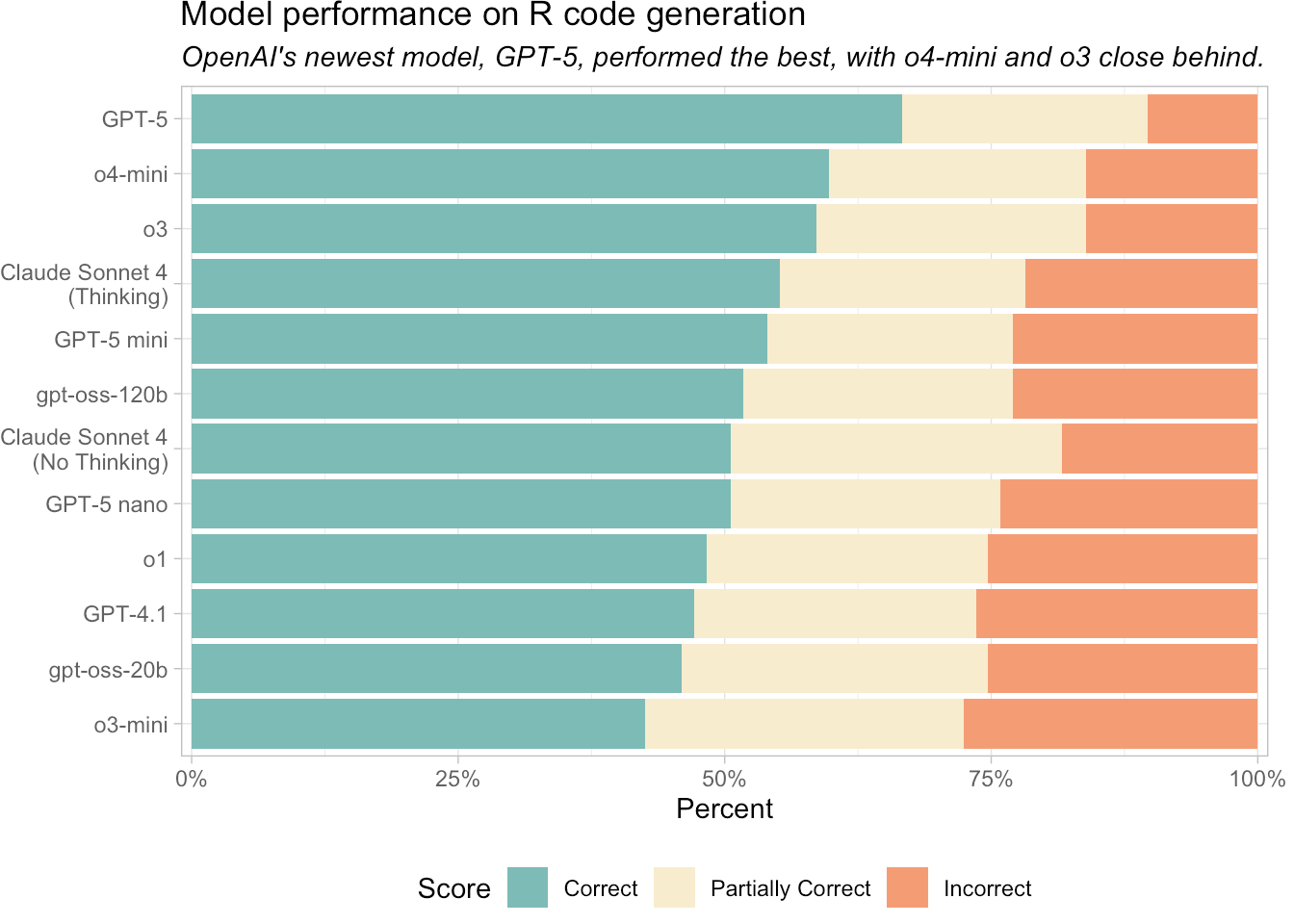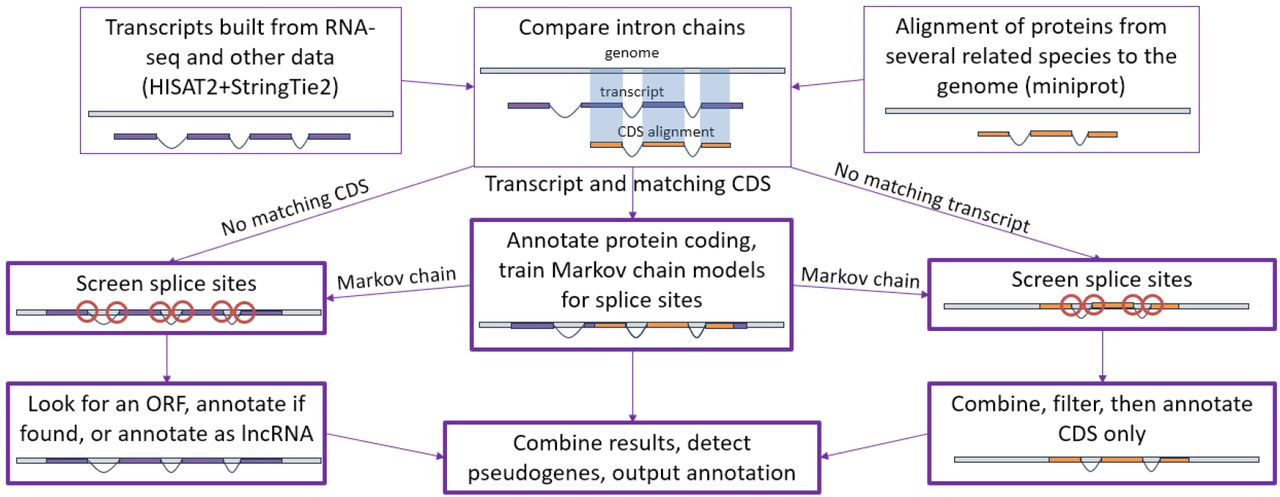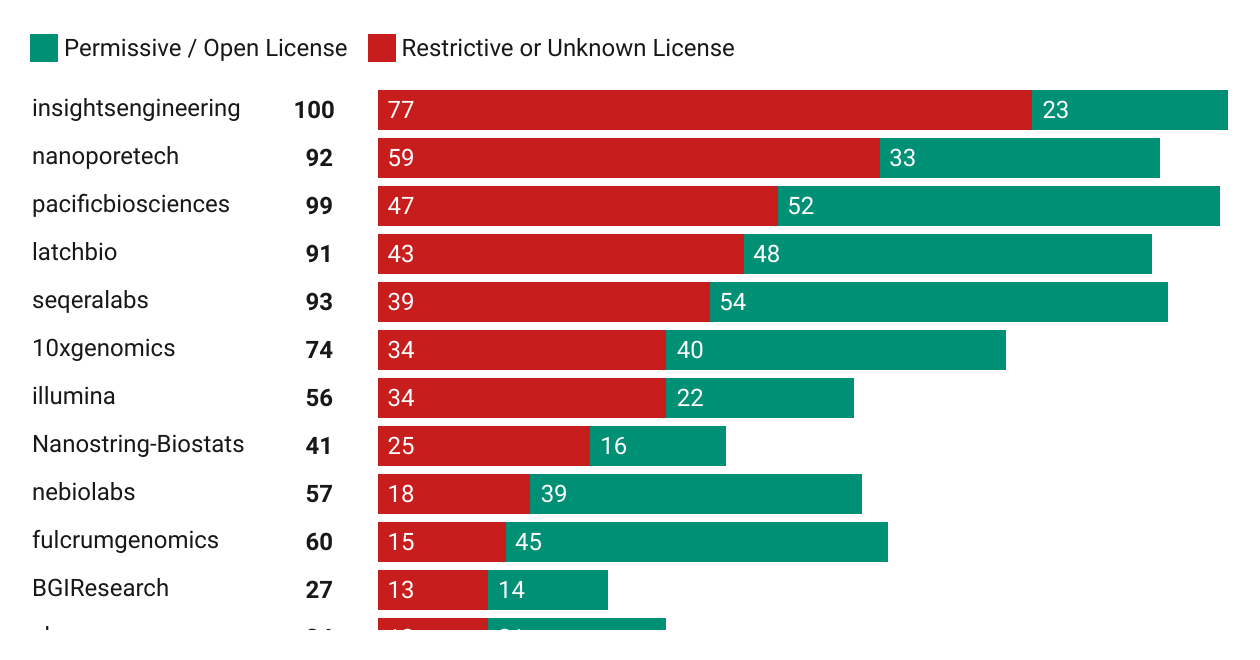
I saw this Tweet a few days ago from Jeremy Leipzig counting the number of GitHub repositories that sequencing-related companies have on their organization page, but was immediately curious how many of these had an open-source license versus a restrictive or unknown license. I tried to one-shot this with GPT-5 given a screenshot of Jeremy’s Tweet and a few instructions. It got me 90% of the way but I had to make a few tweaks here and there.