This blog post is a follow-up to a post in September (Fenner 2022a), where I announced that I had started working on an archive for scholarly blog posts based on the InvenioRDM open-source repository software.

This blog post is a follow-up to a post in September (Fenner 2022a), where I announced that I had started working on an archive for scholarly blog posts based on the InvenioRDM open-source repository software.
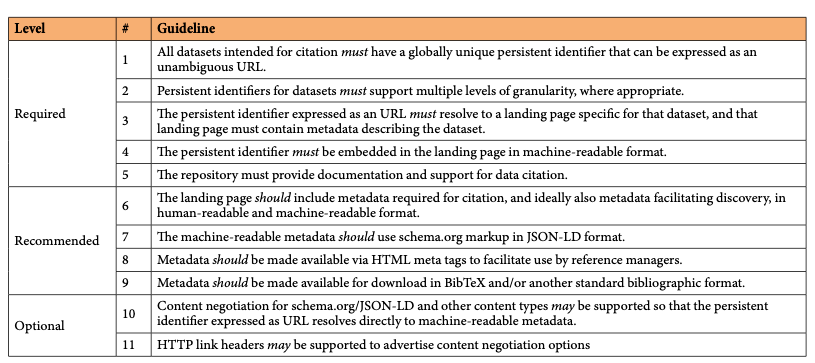
AbstractThis article presents a practical roadmap for scholarly data repositories to implement data citation in accordance with the Joint Declaration of Data Citation Principles, a synopsis and harmonization of the recommendations of major science policy bodies. The roadmap was developed by the Repositories Expert Group, as part of the Data Citation Implementation Pilot (DCIP) project, an initiative of FORCE11.org and the NIH-funded BioCADDIE (https://biocaddie.org) project. The roadmap makes 11 specific recommendations, grouped into three phases of implementation: a) required steps needed to support the Joint Declaration of Data Citation Principles, b) recommended steps that facilitate article/data publication workflows, and c) optional steps that further improve data citation support provided by data repositories. We describe the early adoption of these recommendations 18 months after they have first been published, looking specifically at implementations of machine-readable metadata on dataset landing pages.
In August 2021 I joined the InvenioRDM project to help develop and host a modern repository platform for scholarly content. Things didn't exactly go as planned at the beginning of 2022, and I spent five months in the hospital with serious personal health issues. Since returning home in early June, my health has improved considerably, and in September I was able to slowly start working again.
In January I woke up one day and couldn't move my right arm and leg properly. As I have high blood pressure, and my father had a stroke the exact day seven years ago, I worried that I might have a stroke and went to the emergency room of the local university hospital.
This blog since earlier this month is no longer using a JAMStack setup but a regular Ghost setup using Ghost Pro for hosting. The primary driver were the new native search and native comments, but I needed to do a little bit of work to keep the DOI registration working. This is done now, and an added benefit is that DOI registration is now straightforward for any blog that uses Ghost as a platform.
Almost exactly two years ago, <strong> CrossRef </strong> invited a number of people to discuss unique identifiers for researchers ( <strong> CrossRef Author ID meeting </strong> ). One year ago Thomson Reuters launched <strong> ResearcherID </strong> ( <strong> Thomson Scientific launches ResearcherID to uniquely identify authors </strong> ). And two months ago Phil Bourne and Lynn Fink wrote about this topic in a <strong> PLoS Computational </strong>
Ruby gem and command-line utility for conversion of DOI metadata from and to different metadata formats, including schema.org.