One of the things that didn't make last week's deadline for launching BioNames was the inclusion of phylogenies.

One of the things that didn't make last week's deadline for launching BioNames was the inclusion of phylogenies.
I'm working on displaying OCR text from BHL using SVG, and these are just some quick notes on font size. Specifically how SVG font size corresponds to the size of letters, and how you work out what point size was used to print text on a BHL page. SVG font-size corresponds to the EM square of the font.

Say what you will about Elsevier, they are certainly exploring ways to re-imagine the scientific article. In a comment on an earlier post Fabian Schreiber pointed out that Elsevier have released an app to display phylogenies in articles they publish. The app is based on jsPhyloSVGand is described here.
Following on from the SVG experiments I've started to put some of the Javascript code for displaying phylogenies on Github. Not a repository yet, but as gists, little snippets of code. Mike Bostock has created http://bl.ocks.org/ which makes it possible to host gists as working examples, so you can play with the code "live". The first gist takes a Newick tree, parses it and displays a tree.

Inspired by a comment on my post Visualising edit history of a Wikipedia page, the code I use to make history flow diagrams like the one below is now in GitHub at https://github.com/rdmpage/wikihistoryflow. There is also a live version at http://iphylo.org/~rpage/wikihistoryflow.

I'm taking a virtual part in Mendeley's Hack4Knowledge event. I'm using this a chance to explore some ideas about building novel interfaces to bibliographic data in Mendeley. One idea is to display a user's entire library in one screen. I think the user interfaces employed by most bibliographic software are too conservative and there some cool things that could be done.

Being in an unusually constructive mood, I've spent the last couple of days playing with the TreeBASE II API, in an effort to find out how hard it would be to replace TreeBASE's frankly ghastly interface. After some hair pulling and bad language I've got something to work. It's very crude, but gives a glimpse at what can be done.

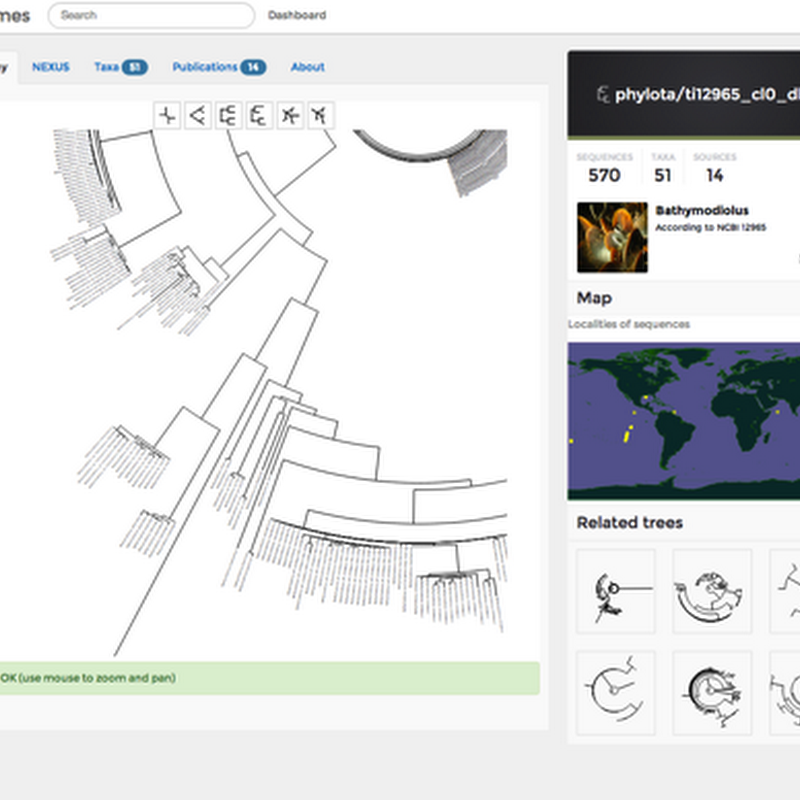
Some serious displacement activity. I'm toying with adding phylogenies to iSpecies, probably sourced from the PhyLoTA browser. This raises the issue of how to display trees on a web page. PhyLoTA itself uses bitmap images, such as this one: but I'd like to avoid bitmaps. I toyed with using SVG, but that has it's own series of issues (it basically has to be served as a separate file). So, I've spent a couple of hours playing with the element.

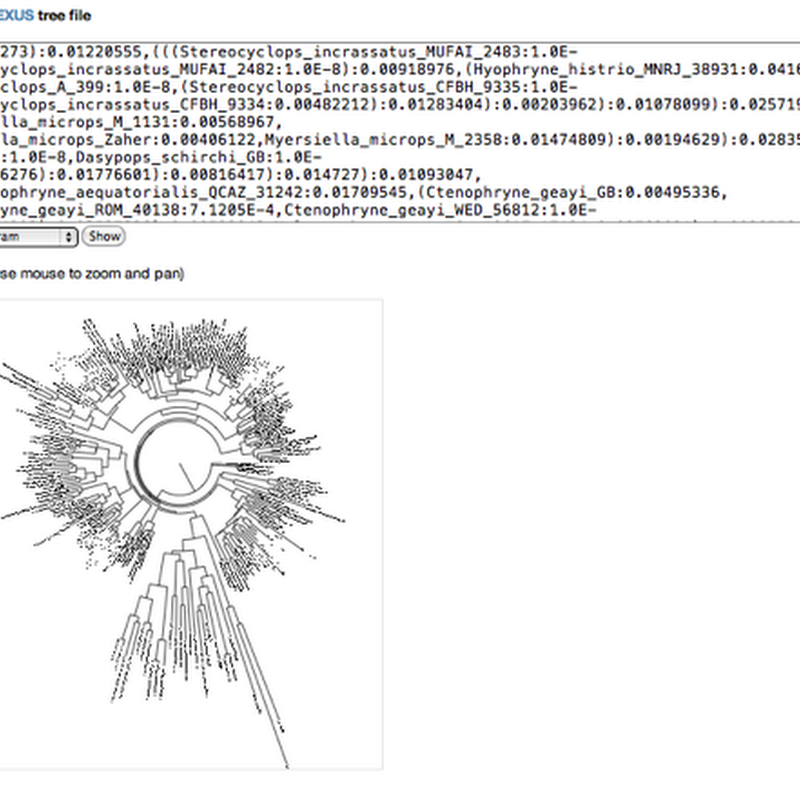
Random half-formed idea time. Thinking about marking up an article (e.g., from PLoS) with a phylogeny (such as the image below, see doi:10.1371/journal.pone.0001109.g001), I keep hitting the fact that existing web-based tree viewers are, in general, crap. Given that a PLoS article is an XML document, it would be great if the tree diagram was itself XML, in particular SVG.

Quick post (really should be doing something else). Reading Jeff Atwood's post Mixing Oil and Water: Authorship in a Wiki World lead me to IBM's wonderful history flow tool to visualise the edit history of a Wikipedia page. There's a nice paper describing history flow (doi:10.1145/985692.985765, free PDF here). Inspired by this I decided to try and implement history flow in PHP and SVG.